Overview of FoldHSphere
Current state-of-the-art deep learning approaches for protein fold recognition learn protein embeddings that improve prediction performance at the fold level. However, there still exists a performance gap at the fold level and the (relatively easier) family level, suggesting that it might be possible to learn an embedding space that better represents the protein folds. In this paper, we propose the FoldHSphere method to learn a better fold embedding space through a two-stage training procedure. We first obtain prototype vectors for each fold class that are maximally separated in hyperspherical space. We then train a neural network by minimizing the angular large margin cosine loss (LMCL) to learn protein embeddings clustered around the corresponding hyperspherical fold prototypes. Our network architectures, ResCNN-GRU and ResCNN-BGRU, process the input protein sequences by applying several residual-convolutional blocks followed by a gated recurrent unit-based recurrent layer. Evaluation results on the LINDAHL dataset indicate that the use of our hyperspherical embeddings effectively bridges the performance gap at the family and fold levels. Furthermore, our FoldHSpherePro ensemble method yields an accuracy of 81.3% at the fold level, outperforming all the state-of-the-art methods. Our methodology is efficient in learning discriminative and fold-representative embeddings for the protein domains. The proposed hyperspherical embeddings are effective at identifying the protein fold class by pairwise comparison, even when amino acid sequence similarities are low.
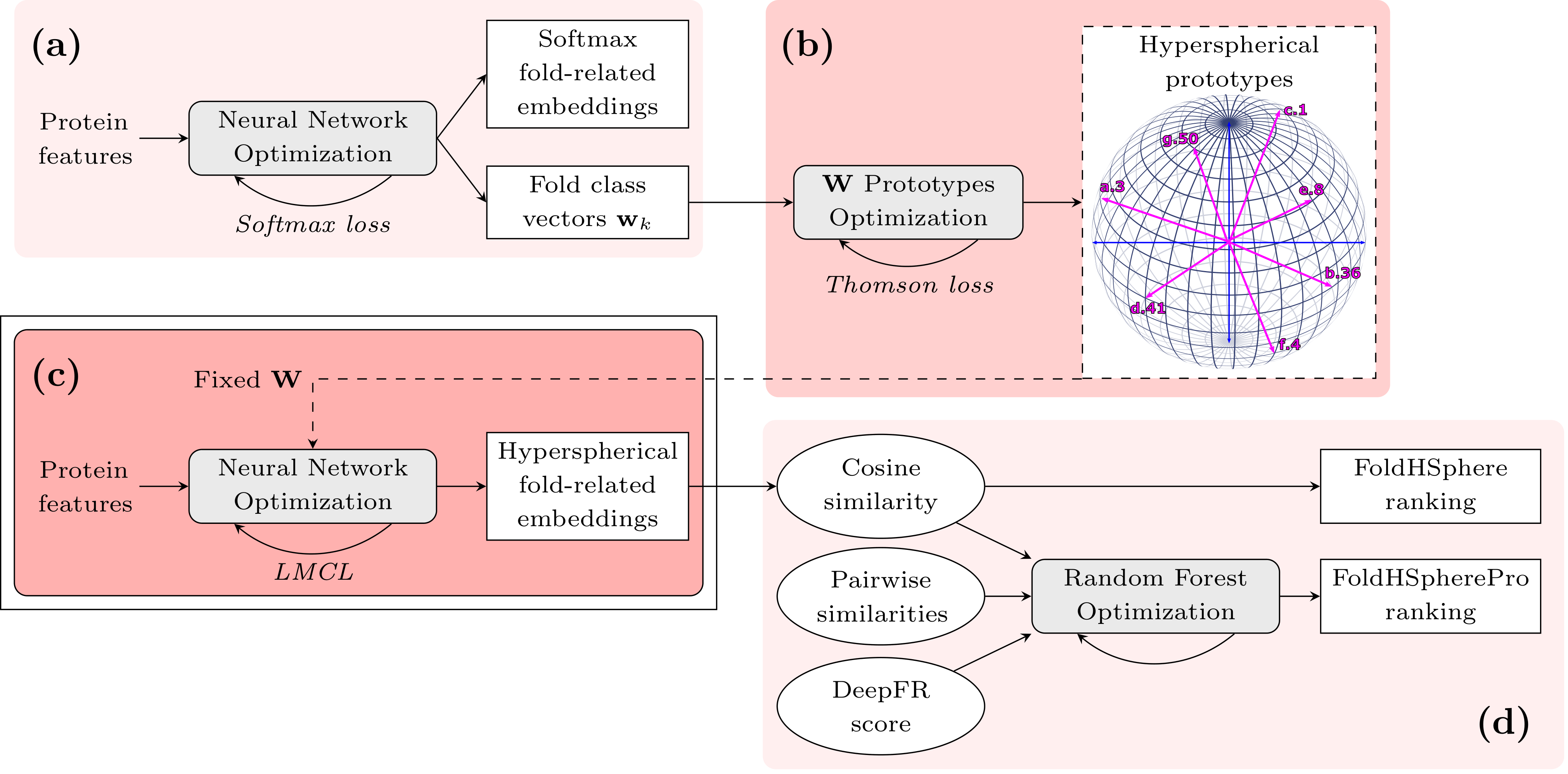
Figure 1. Overview of the FoldHSphere approach for protein fold recognition. In the first stage (a) we train a neural network model to map the protein domains into K fold classes using the softmax cross-entropy as loss function. From this trained model, we extract fold class weight vectors wk (k=1,...,K) learned in the last classification layer. (b) We then optimize the position of the wk vectors by our proposed Thomson-based loss, so that they are maximally separated in the angular space. (c) The resulting hyperspherical prototypes are used as a fixed non-trainable classification matrix W in the last layer of the neural network model, which is trained again, but now minimizing the LMCL. The final hyperspherical embeddings are extracted from the fully-connected part of this model. (d) Finally, the cosine similarity is computed between each two embeddings and a template ranking is performed for each query protein domain (FoldHSphere method). Moreover, template ranking is further improved by using enhanced scores provided by a random forest model trained with additional similarity measures as inputs (FoldHSpherePro method).
Paper access
Villegas-Morcillo, A., Sanchez, V. & Gomez, A.M. FoldHSphere: deep hyperspherical embeddings for protein fold recognition. BMC Bioinformatics 22, 490 (2021). DOI: 10.1186/s12859-021-04419-7
GitHub
Link to GitHub code: https://github.com/amelvim/FoldHSphere
Downloadable data
Source code, data, features, and trained models (Updated 04-05-2021)
Contact: Amelia Villegas-Morcillo