Overview
The identification of the protein fold class is a challenging problem in structural biology. Recent computational methods for fold prediction leverage deep learning techniques to extract protein fold-representative embeddings mainly using evolutionary information in the form of multiple sequence alignment (MSA) as input source. In contrast, protein language models (LM) have reshaped the field thanks to their ability to learn efficient protein representations (protein-LM embeddings) from purely sequential information in a self-supervised manner. In this paper, we analyze a framework for protein fold prediction using pre-trained protein-LM embeddings as input to several fine-tuning neural network models which are supervisedly trained with fold labels. In particular, we compare the performance of six protein-LM embeddings: the LSTM-based UniRep and SeqVec, and the transformer-based ESM-1b, ESM-MSA, ProtBERT, and ProtT5; as well as three neural networks: Multi-Layer Perceptron (MLP), ResCNN-BGRU (RBG), and Light-Attention (LAT). We separately evaluated the pairwise fold recognition (PFR) and direct fold classification (DFC) tasks on well-known benchmark datasets. The results indicate that the combination of transformer-based embeddings, particularly those obtained at amino acid-level, with the RBG and LAT fine-tuning models performs remarkably well in both tasks. To further increase prediction accuracy, we propose several ensemble strategies for PFR and DFC, which provide a significant performance boost over the current state-of-the-art results. All this suggests that moving from traditional protein representations to protein-LM embeddings is a very promising approach to protein fold-related tasks.
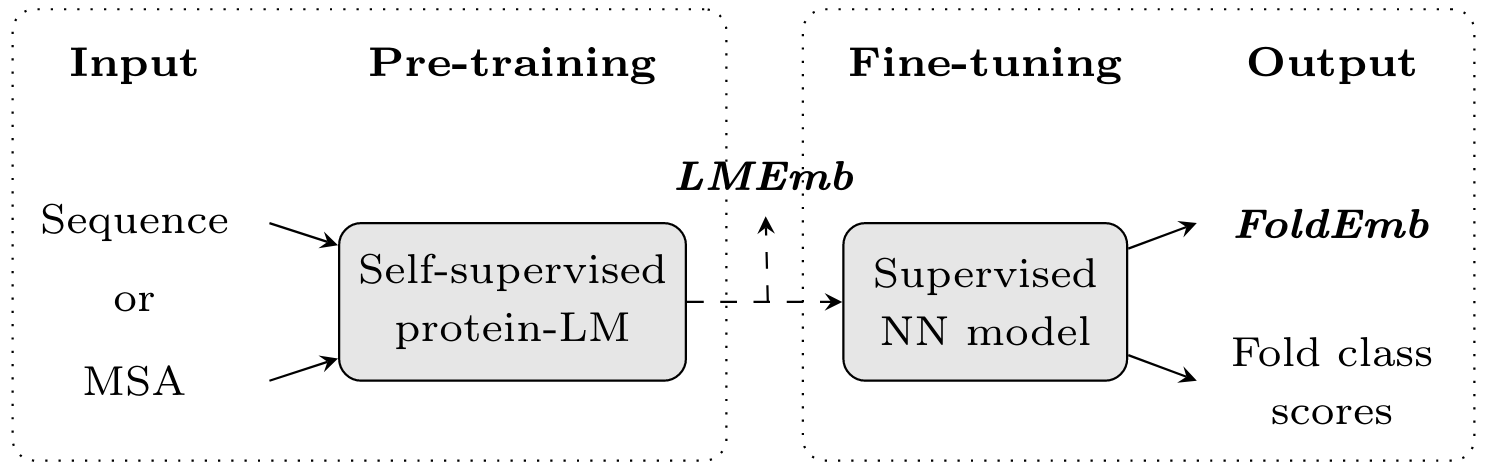
Figure 1. Overview of our approach for protein fold prediction. First, we extract a protein embedding representation from the amino acid sequence or multiple sequence alignment (MSA) using protein language models (protein-LMs) which have been pre-trained in a self-supervised manner (i.e. using the input sequential information itself). As a result, we obtain a protein-LM embedding (LMEmb) of size LxF, where L is the length of the protein sequence and F is the size of the amino acid-level embedding. Then, we fine-tune this embedding through a neural network (NN) model that is trained, in a supervised manner, to map the input protein into K fold classes. The outputs of this model are, on the one hand, a fold-representative embedding of the protein (FoldEmb with fixed-size 512), used to perform the pairwise fold recognition (PFR) task; and, on the other hand, the scores for each fold class, used in the direct fold classification (DFC) task.
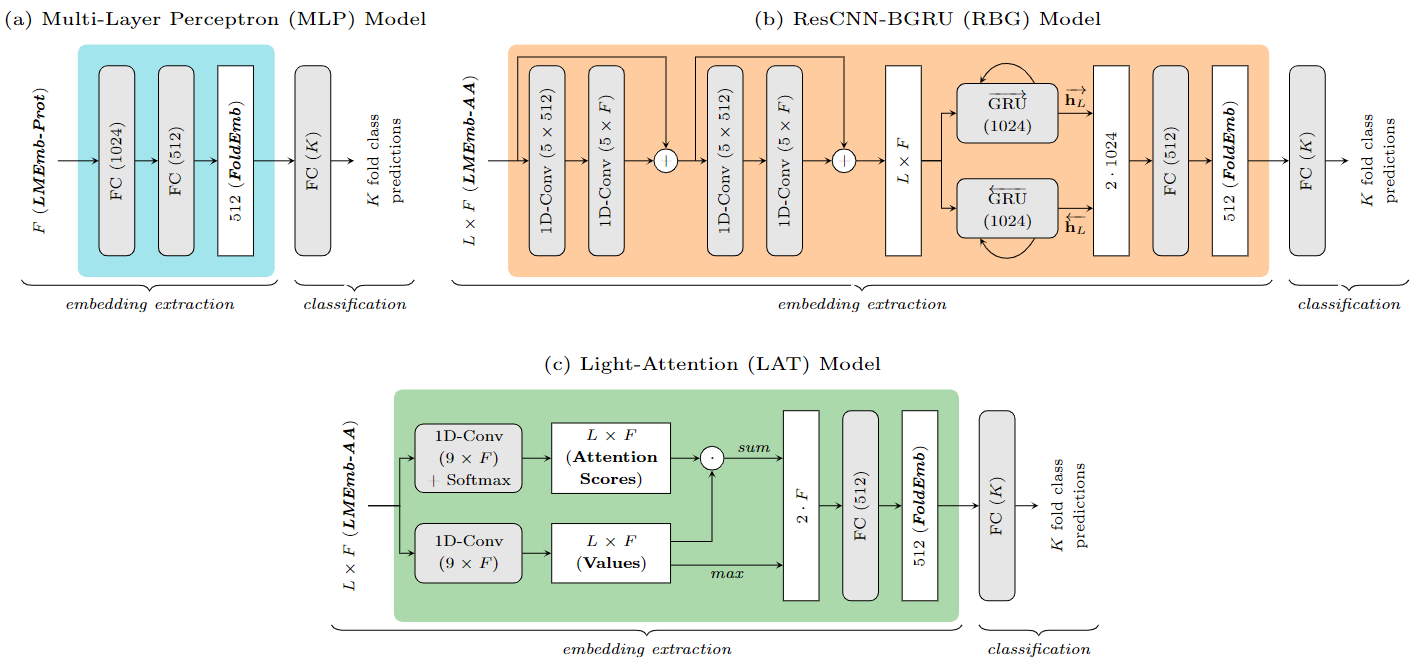
Figure 2. Neural network models used to fine-tune the protein-LM embeddings (LMEmb) to fold-representative embeddings (FoldEmb), as well as to perform direct fold classification. Three neural architectures are used as embedding extractors (identified with three distinct background colors). (a) The Multi-Layer Perceptron (MLP) model processes the protein-level embeddings (LMEmb-Prot) through two fully-connected (FC) layers. (b) The ResCNN-BGRU (RBG) model (Villegas-Morcillo et al. 2021) processes the amino acid-level embeddings (LMEmb-AA) through two residual-convolutional blocks, a bidirectional gated recurrent unit (GRU) layer, and an FC layer. (c) The Light-Attention (LAT) model, adapted from (Stark et al. 2021), also processes LMEmb-AA through an attention mechanism followed by an FC layer.
Paper access
A. Villegas-Morcillo, A.M. Gomez, and V. Sanchez, "An Analysis of Protein Language Model Embeddings for Fold Prediction," Briefings in Bioinformatics, bbac142 (2022). DOI: 10.1093/bib/bbac142
GitHub
Link to GitHub code: https://github.com/amelvim/FoldEmbeddings
Downloadable data
Data, LM embeddings, and trained models (Updated 20-01-2022)
Contact: Amelia Villegas-Morcillo