Protein Fold Recognition from Sequences using Convolutional and Recurrent Neural Networks
The identification of a protein fold type from its amino acid sequence provides important insights about the protein 3D structure. In this paper, we propose a deep learning architecture that can process protein residue-level features to address the protein fold recognition task. Our neural network model combines 1D-convolutional layers with gated recurrent unit (GRU) layers. The GRU cells, as recurrent layers, cope with the processing issues associated to the highly variable protein sequence lengths and so extract a fold-related embedding of fixed size for each protein domain. These embeddings are then used to perform the pairwise fold recognition task, which is based on transferring the fold type of the most similar template structure. We compare our model with several template-based and deep learning-based methods from the state-of-the-art. The evaluation results over the well-known LINDAHL and SCOP_TEST sets, along with a proposed LINDAHL test set updated to SCOP 1.75, show that our embeddings perform significantly better than these methods, specially at the fold level.
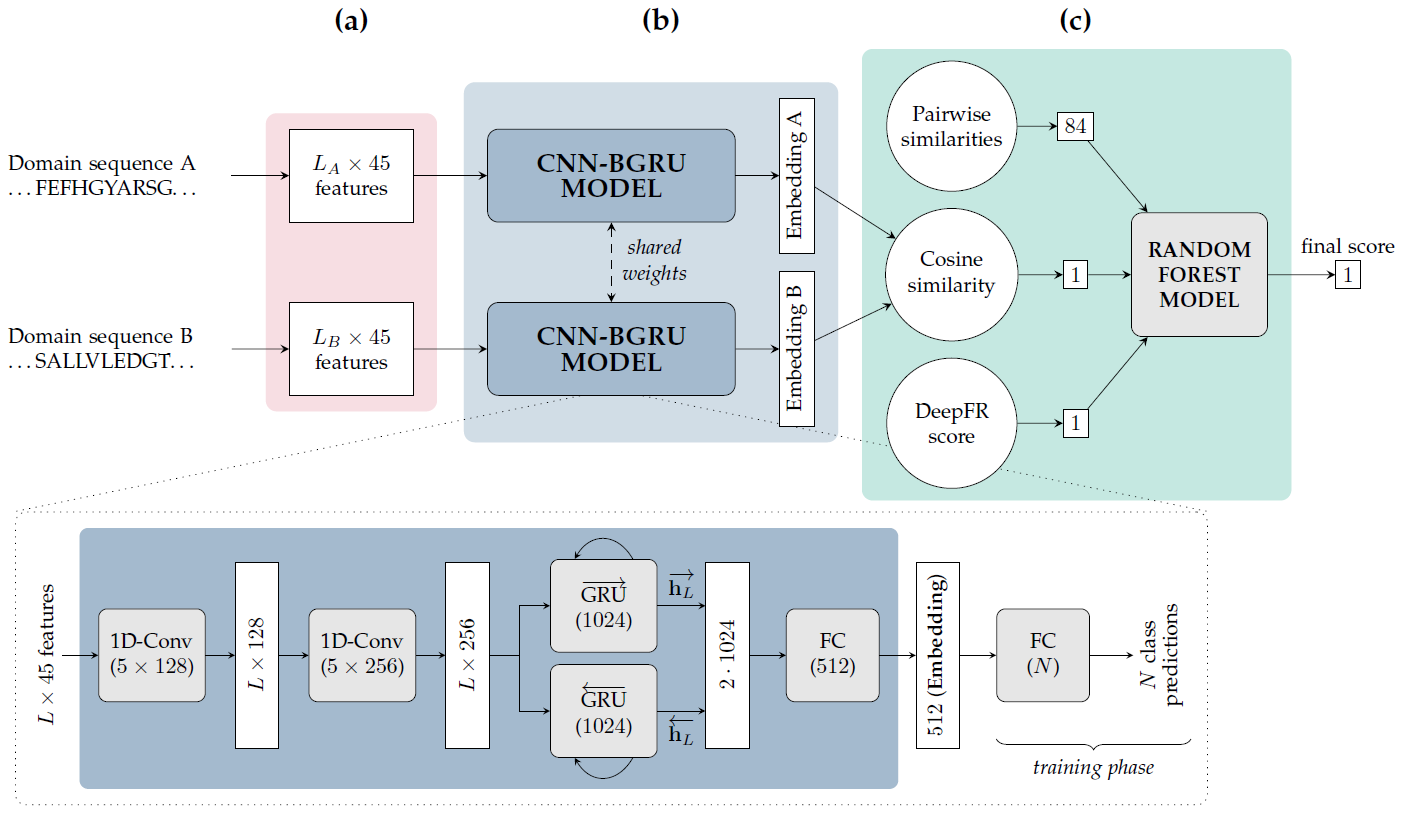
Figure 1. The proposed fold recognition approach to obtain a fold similarity score for two protein domains. (a) First, 45 input features are extracted for each amino acid in each of the two domain sequences. (b) The resulting Lx45 residue-level features are passed through the CNN-BGRU model, previously trained to map protein sequences into fold classes. The model architecture (bottom part) consists of two one-dimensional convolutional layers, a bidirectional gated recurrent unit (GRU) layer and two fully-connected (FC) layers. The CNN-GRU model is very similar but with a unidirectional GRU instead and hence an output of size 1024. The two input protein domains are processed independently by the same network model (i.e. with identical trained weights). This model is then used to extract a fold-related embedding vector for each one (from the 512-dimensional output of the first FC layer). (c) The cosine similarity distance is computed between the two embeddings, which is concatenated to other similarity measures in a vector to obtain the final fold similarity score using a random forest model.
Paper access
Villegas-Morcillo, A., Gomez, A.M., Morales-Cordovilla, J.A., and Sanchez, V. Protein Fold Recognition from Sequences using Convolutional and Recurrent Neural Networks. IEEE/ACM Transactions on Computational Biology and Bioinformatics (2020). DOI: 10.1109/TCBB.2020.3012732
Downloadable data
Data, Features, Models and Code (Updated 18-02-2020)
Contact: Amelia Villegas-Morcillo